Using DNA barcoding to genetically identify inbred Drosophila lines
Working with a large collection of Drosophila melanogaster strains takes large amounts of manual work. First, to ensure a stable growth of the different strains. Second, to generate enough sample material for experiments. Because of this chances are that by mistake sometimes strains are mis-labeled or get mixed up. These small mistakes of-course negatively influence all analysis further downstream.
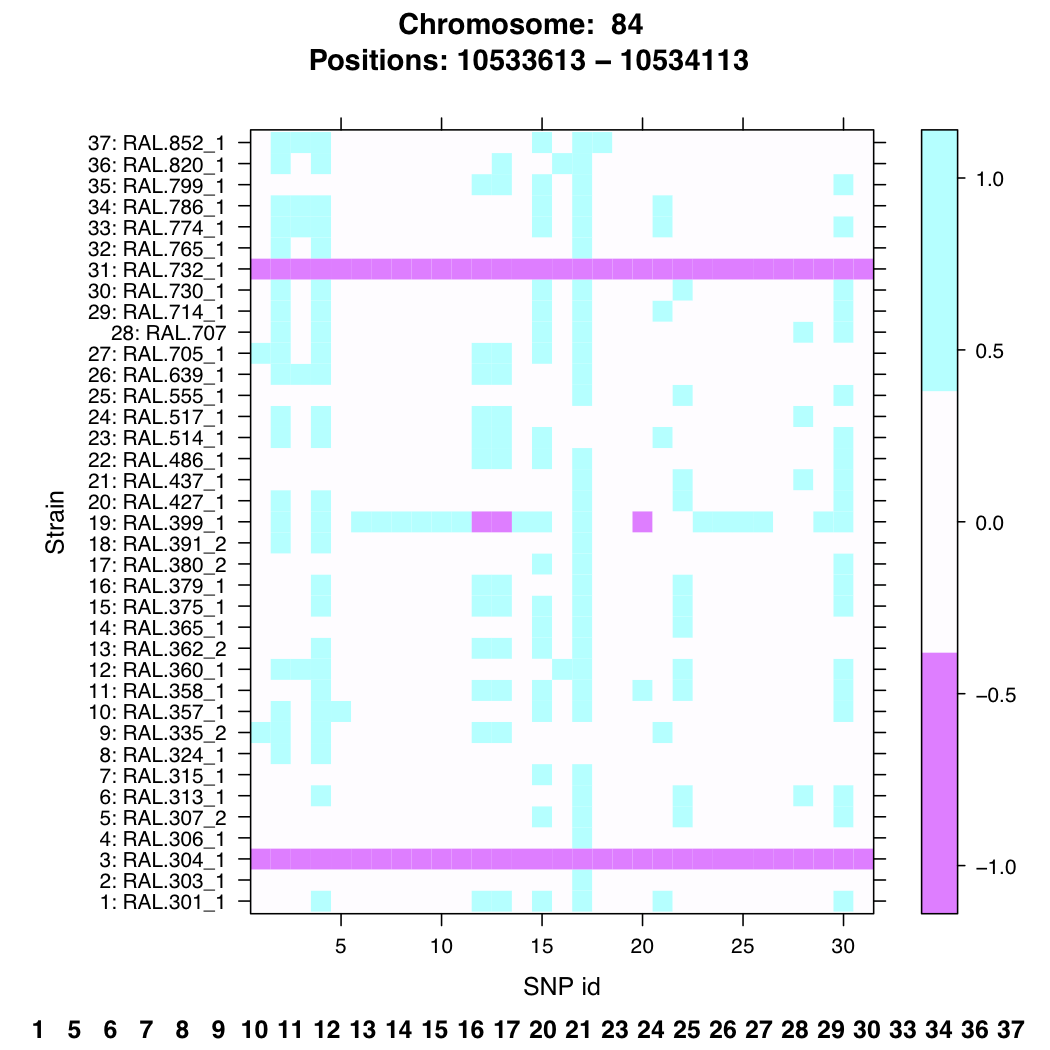
In meta-genomics projects they use known polymorphic sites (ranging from microsatellites to SNPs) to identify what organisms are inside a sample. Now that within the Drosophila Genetic Reference Panel (DGRP) all inbred strains are fully sequenced we can use the polymorphic sites (in our case SNPs) to identify each strain uniquely using the same barcoding idea. I developed several functions in R that help selecting regions to be targeted using cheap old-fashioned RT-PCR to identify all of the strains uniquely without having to construct unique primers for each individual strain.
In the example below we selected a region that, when sequenced, is able to identify 28 unique strains using the 31 SNPs inside this specific region. By selecting several of this regions you can identify every strain with a high confidence.