De-novo sequence assembly of high coverage genomes
As part of my PhD studies I was working on some individuals from the Drosophila Genetic Reference Panel (DGRP) that are sequenced with high coverage. For 2 strains (RAL 375 and 852) an average sequence coverage of 25X sequence data is available making things like de-novo sequence assemblies possible. Using the Velvet we constructed several de-novo assemblies for both individuals (n50 of 40k and 50k) and studied these assemblies to check for sequence divergency and potential large structural variance.
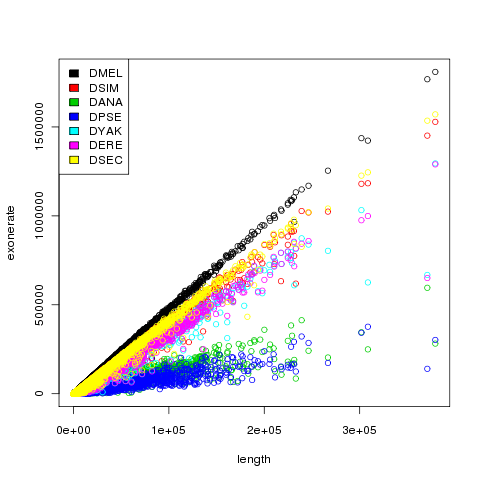
We took the contigs constructed by velvet for each of the individuals and aligned them against different reference genomes of Drosophila and some close related species using Exonerate. By plotting the length versus exonerate score (as a measurement of sequence similarity of contigs towards the reference genomes) it becomes clear that contigs of our de-novo sequence assemblies show the largest sequence similarity with the Drosophila melanogaster reference genome, something that was of-course expected. This is a positive result that strengthens our believe into that you can use sequence assemblers like Velvet to re-construct unknown genomes when sufficient sequence coverage is available.
One contig is showing a more interesting result that we haven’t been able to explain sufficient enough. This one contig is showing a much lower exonerate score than expected based on the contig length (exonerate score is scaling linear with contig length normally). This could indicate that in this specific contig something biological relevant is happening or that in this specific contig Velvet made a mistake. With the current data and coverage for that area we cannot conclude any of these results.