The influence of protein dynamics on structural alignments
Minor project at the The Centre for Integrative Bioinformatics, Vrije Universiteit, Amsterdam, The Netherlands. Under supervision of Prof Heringa, Dr. Feenstra and Dr. Pirovano.
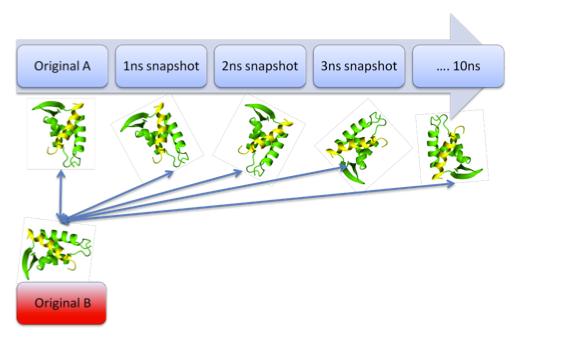
Current structural alignment methods don’t consider structural variance. By modelling protein dynamics on HOMSTRAD homologous proteins (with GROMACS) a new set of structural variant protein structures was created that respond to different “normal” stages of a protein structure. Dalilite structural alignments were made made and scored using sum-of-pairs. By comparing the sum-of-pairs score obtained with Dalilite and the RMSD divergence scores from our simulations we gained insight in how well structural alignment tools can deal with different sort of normal structural variance.
Part of my work has been published in the following publication: The meaning of alignment: lessons from structural diversity